ri bio api search tool
Mar 2021 - Aug 2021
Background
During the spring of 2021, I joined Pangea, a freelancer network made up of college students. I was connected with RI Bio, a group aiming to improve the life sciences industry in Rhode Island.
Prior to the development of this tool, researchers looked up the same term in 4 different databases to see what existed so far. RI Bio was looking for a more efficient search for their use cases - they wanted to receive results from all databases in one search, and be able to see titles, authors, and other general information. From this basic information, if something was intriguing, they could then go look it up in the specific database.
In additional to basic search results, there were some analyses that could give good signals on the search term.
Field Expert: Who’s name pops up the most across papers, patents, grants, and trials? (This may potentially be a good person to reach out to)
Patent Similarity: Without needing to read every patent, what patents are focused on similar ideas and technologies?
Note: the researchers/users of this tool did not have a CS technical background, so the setup and usage of the tool could not be too technically involved.
Implementation
Django was used to build an API which would send requests to each of the database’s APIs, compile and analyze the data, and output a excel file with multiple sheets.
The full source code can be found here, and there will be links to specific files below.
Quick Overview
Implementation of this was a little tedious but fairly straightforward - it involved following the API docs for each database, writing requests in the correct format, and displaying relevant information in an excel sheet. The four databases queried from are shown below. They link to the source code used to connect to the specific API.
-
ClinicalTrials.gov - clinical trials
-
NIH Federal Reporter - projects granted federal money
-
Lens Scholarly Search - scholarly publications
-
Lens Patent Search - patents
Analysis - Field Expert
First, we would pull all names (authors, primary investigators, inventors, etc.).
All the name patterns were slightly different (Last, First vs First Middle Last), but after transforming all the names into the same pattern, this was also fairly straightforward - a count was associated with each name, and the names were shown on a list in descending order, alongside the titles of the paper/patent/grant/trial they appeared in.
The source code can be found here. The “translator” dictionary at the beginning tracks the columns the names appear in, and the indices of the first and last name (databases that stored names as Last First would have indices 1, 0 in this case). The Lens databases are commented out because in the last iteration of this code, the Lens API key had expired.
Analysis - Patent Similarity
The process for comparing all patents is as follows:
-
For each patent, get the claims (what does this patent cover), and ignore common words (a, the, is, etc.).
-
Use set similarity search, comparing all possible pairs, to see which pairs of patents have the highest similarity.
Note - if you look at the source code for this section, you will see that the TF-IDF for each word in each patent was calculated. This was done in case a future version included an improved comparison which accounted for not only the words, but how unique and important they were.
Final State
Unfortunately, it is difficult to show a proper demo for this tool - the Lens API key has expired, and the NIH Federal Reporter API was retired.
However, I have some screenshots and past downloads to give some idea of what it looked like while functional.
Following the instructions on the Github project page would display the page shown on the right:
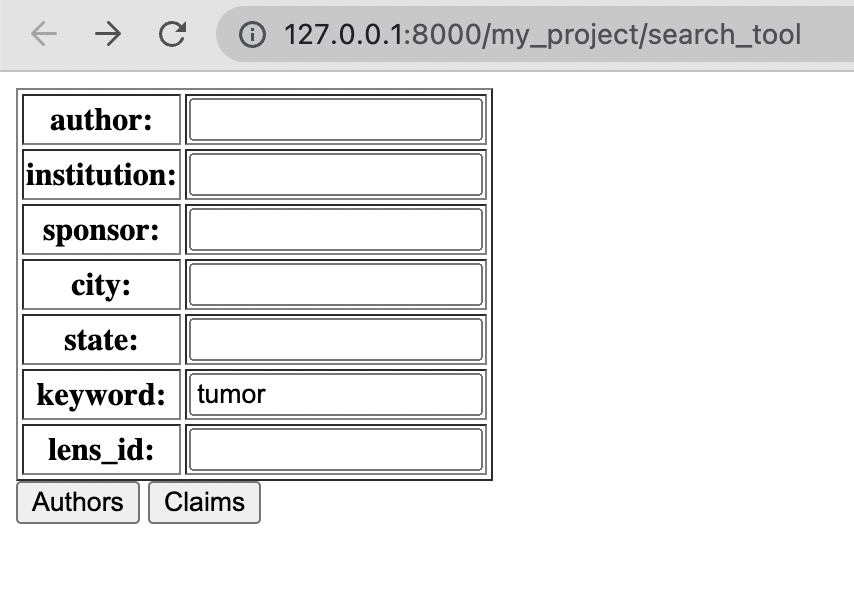
The user would primarily be interacting with the “keyword” section. They could filter results by the author, location, etc. Where applicable, these filters would be inserted into the API request.
The Lens ID was for convenience - if a user had a specific Lens ID (a specific scholarly article/patent) they wanted to look up within this tool, they could do so.
After entering different parts of the query, the user could choose to see an author analysis (“Authors”) of the query or a patent analysis (“Claims”). The two analyses were separated because the patent analysis was significantly slower, so the user didn’t need to wait for it unless they needed to.
Click below to see the results of looking up certain terms in the tool.
- Nitinol Paddle Lead (no filters)
- This term yielded no results from the Clinical Trials database or the NIH database.
- This was a “Claims” search, and the analyses tab displays similar patents. Index1/Index2 in the Claims tab map to patent’s index in the lens_p_results tab, so it is easy to get more information.